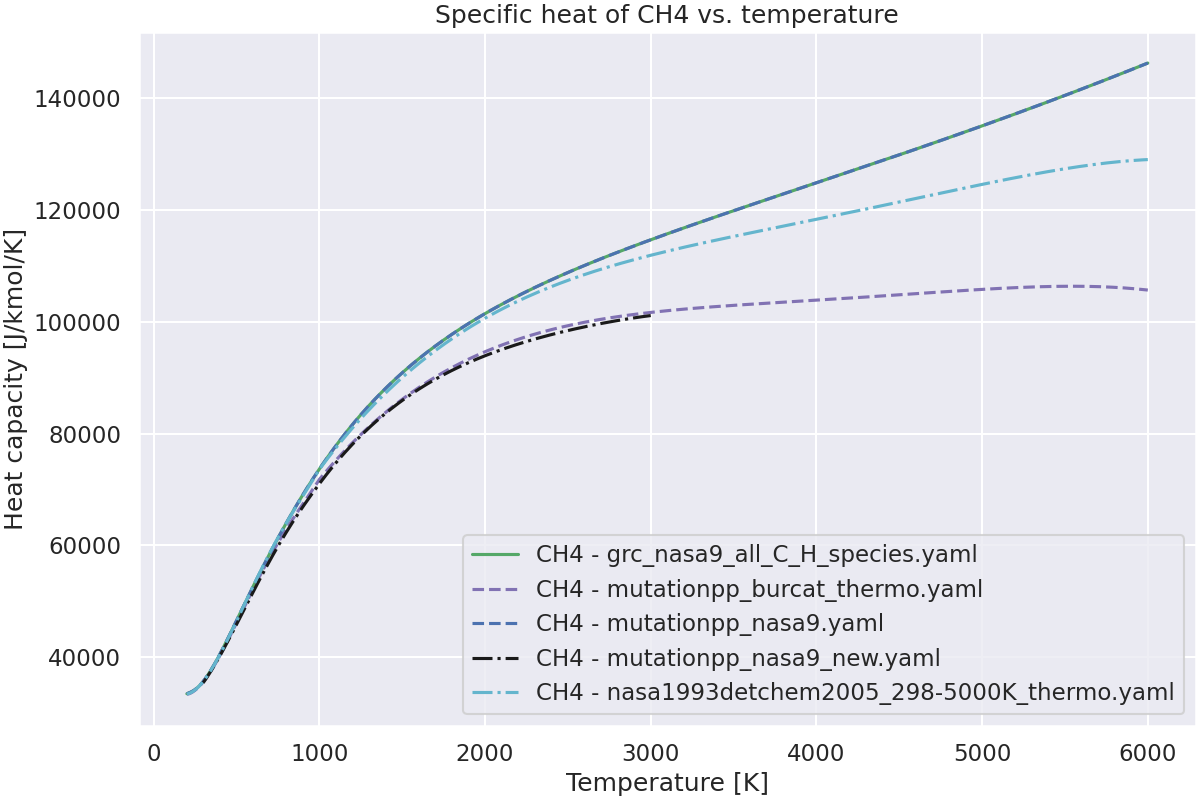
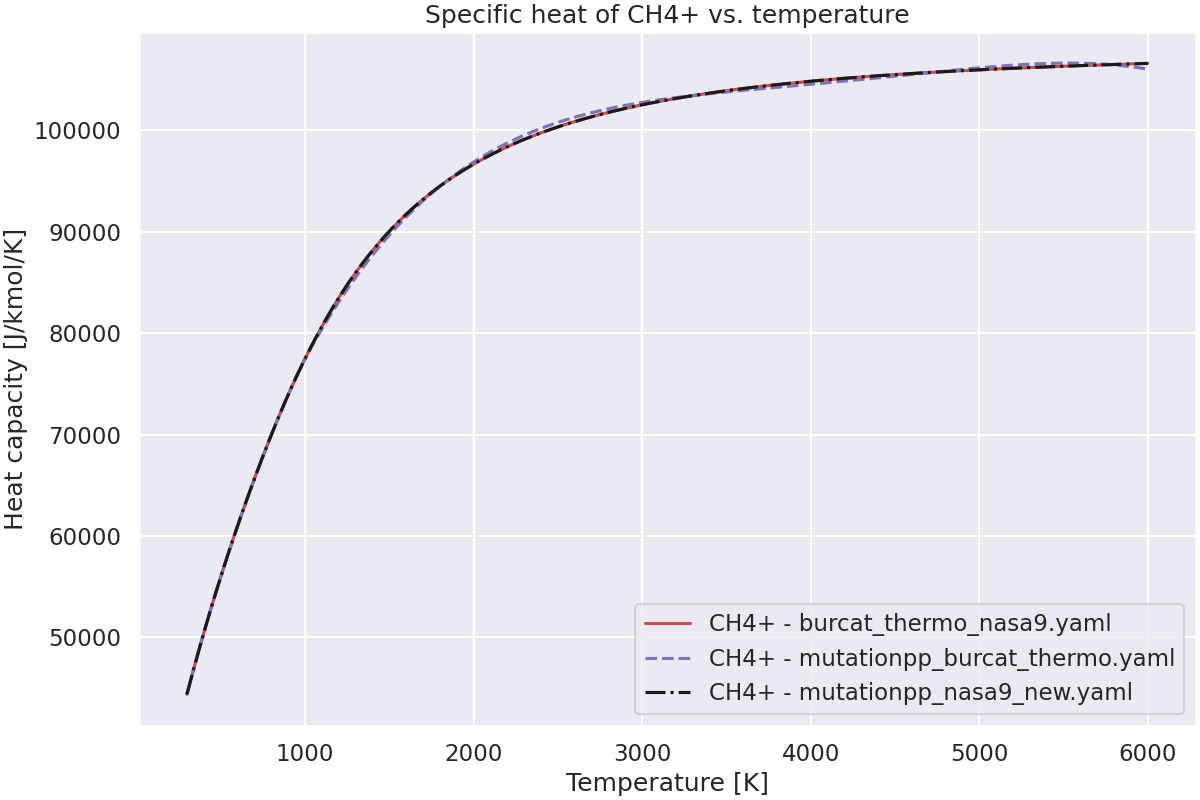
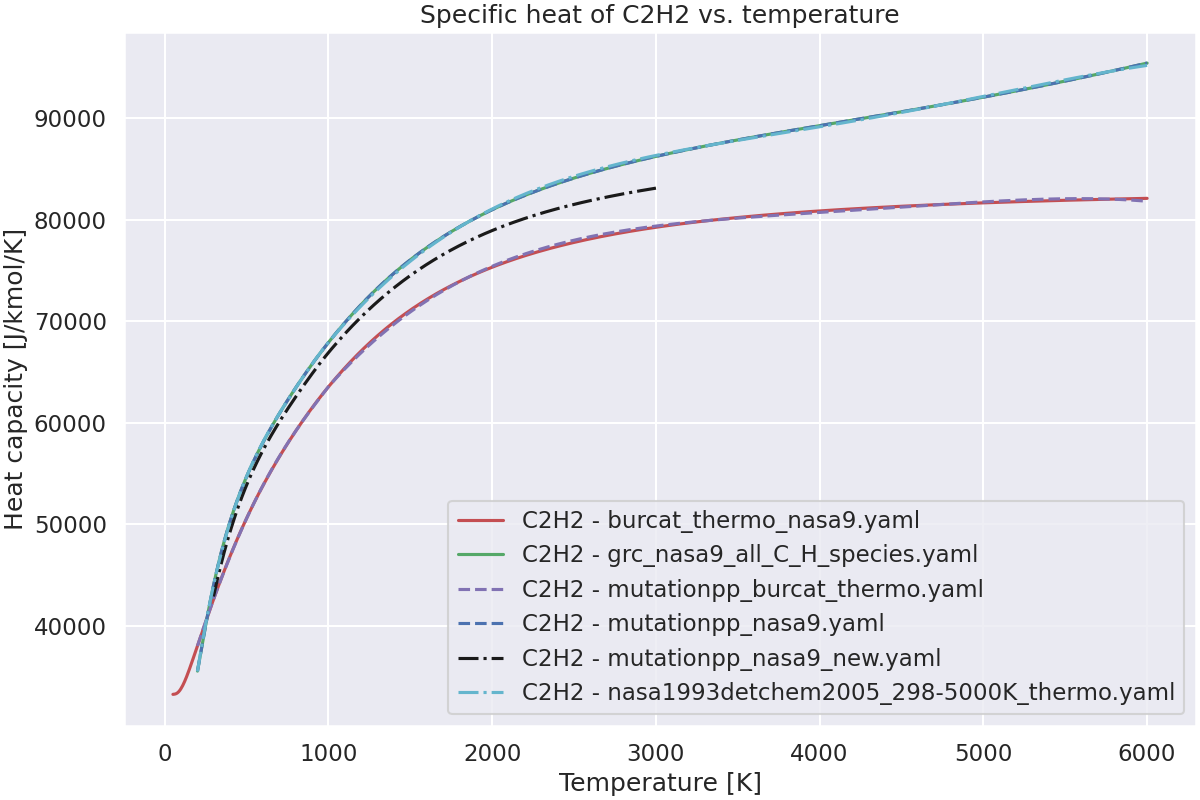
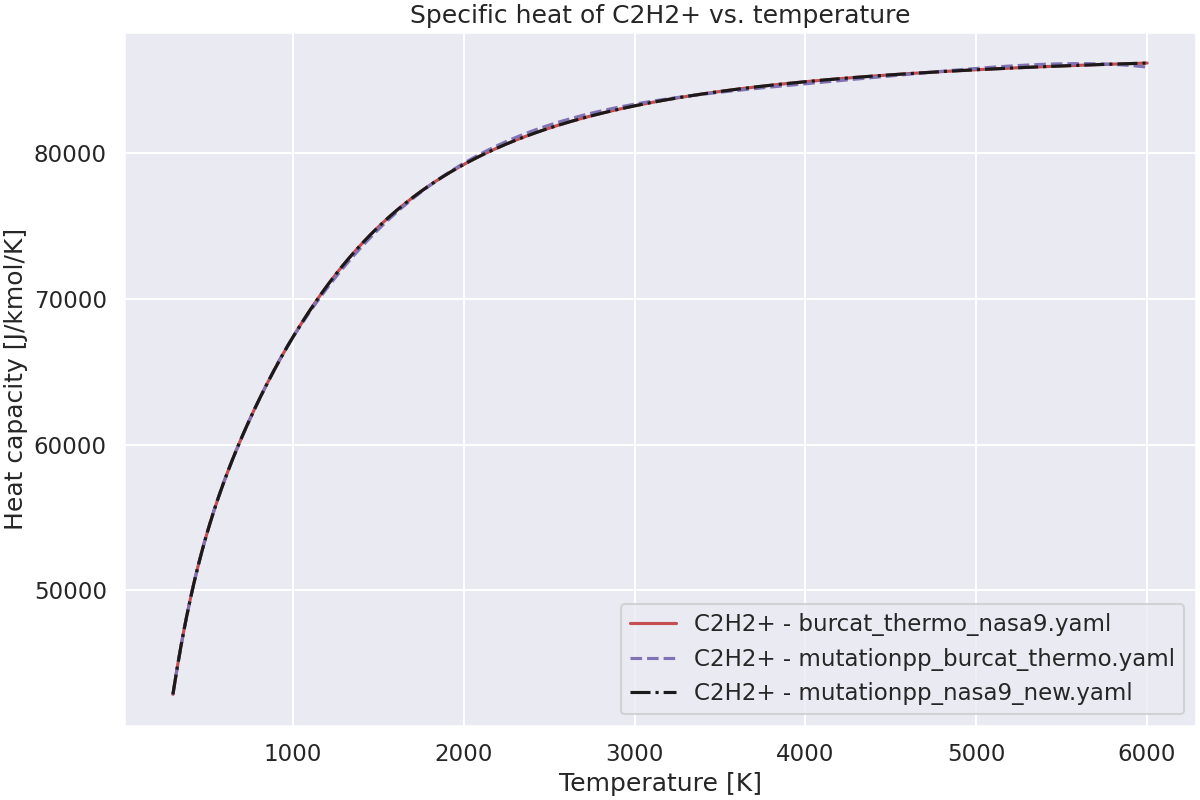
—
Plot specific heat vs. temperature for different species and thermo data.#
This example demonstrates how to plot the specific heat of different species vs. temperature for different thermo data files.
The specific heat of a species is a function of temperature. The specific heat is the amount of heat required to raise the temperature of a unit mass of a substance by one degree Kelvin.
Here, the specific heat is in J/kmol/K, and the temperature is in Kelvin.
# This is an option for the online documentation, so that each image is displayed separately.
# sphinx_gallery_multi_image = "single"
Import the required libraries.#
import cantera as ct
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from rizer.misc.utils import get_path_to_data
sns.set_theme("talk")
Define the thermo data files to load.#
# Dictionary to store thermo data files.
# Key: name of the thermo data file.
# Value: color and linestyle, to use in the plot.
thermo_data: dict[str, str] = {
"burcat_thermo_nasa9.yaml": "r",
"grc_nasa9_all_C_H_species.yaml": "g",
"mutationpp_burcat_thermo.yaml": "m--",
"mutationpp_nasa9.yaml": "b--",
"mutationpp_nasa9_new.yaml": "k-.",
"nasa1993detchem2005_298-5000K_thermo.yaml": "c-.",
}
Set the species to plot.#
# Dictionary to store species data.
# Key: species name
# Value: dictionary with:
# key: name of the thermo data
# value: Cantera Species object
species_to_plot: dict[str, dict[str, ct.Species]] = {
"CH4": {},
"CH4+": {},
"C2H2": {},
"C2H2+": {},
"H2": {},
"H2+": {},
}
Load species data from the various mechanisms.#
for thermo in thermo_data:
# Load species data.
path_to_species = get_path_to_data("thermo", thermo)
species_list: list[ct.Species] = ct.Species.list_from_file(str(path_to_species))
# Look up for matching species in the list.
for species_name in species_to_plot:
for specie in species_list:
if specie.name == species_name:
species_to_plot[species_name][thermo] = specie
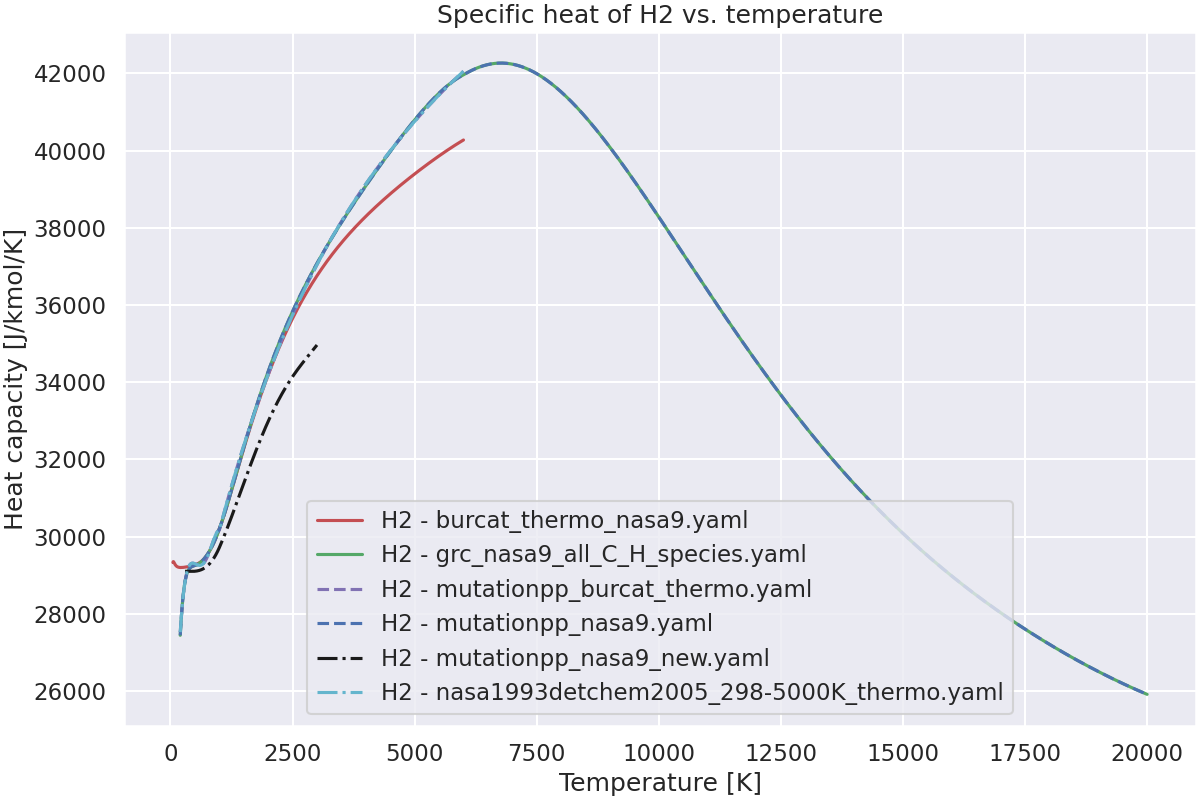
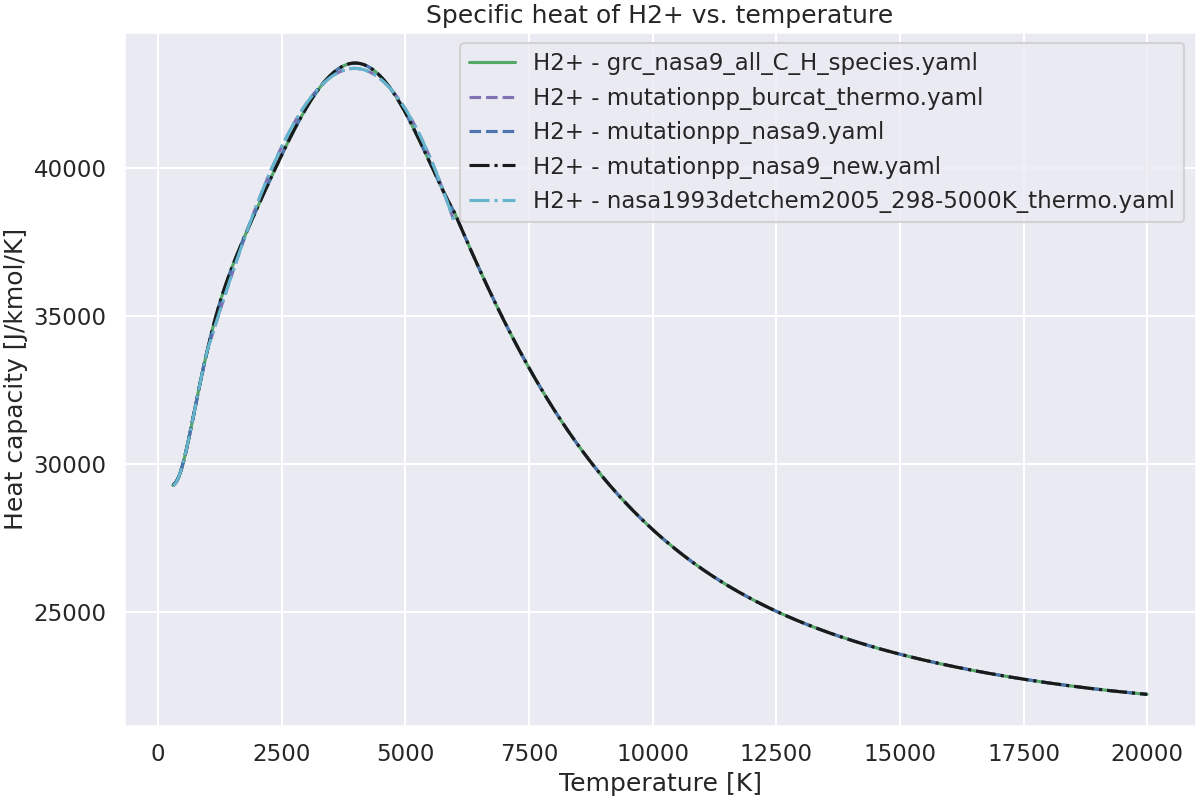
Plot the specific heat of the species vs. temperature.#
# For each species, plot the specific heat vs. temperature.
for species_name in species_to_plot:
fig, ax = plt.subplots(figsize=(12, 8), layout="constrained")
for thermo in thermo_data:
# Check if the species is present in the thermo data.
if thermo not in species_to_plot[species_name]:
print(
f"Species {species_name} not found in {thermo}."
"Check spelling and composition."
)
continue
# Get the species object.
species = species_to_plot[species_name][thermo]
# Define the temperature range.
min_temperature = species.thermo.min_temp
max_temperature = species.thermo.max_temp
temperatures = np.linspace(min_temperature, max_temperature, num=1000) # K
# Compute the specific heat for each temperature.
cp = np.array([species.thermo.cp(T) for T in temperatures]) # J/kmol/K
# Plot the specific heat vs. temperature.
color = thermo_data[thermo]
ax.plot(temperatures, cp, color, label=f"{species_name} - {thermo}")
ax.set_title(f"Specific heat of {species_name} vs. temperature")
# Set the labels and legend.
ax.set_xlabel("Temperature [K]")
ax.set_ylabel("Heat capacity [J/kmol/K]")
# Plot
plt.legend()
plt.show()
Species CH4 not found in burcat_thermo_nasa9.yaml.Check spelling and composition.
Species CH4+ not found in grc_nasa9_all_C_H_species.yaml.Check spelling and composition.
Species CH4+ not found in mutationpp_nasa9.yaml.Check spelling and composition.
Species CH4+ not found in nasa1993detchem2005_298-5000K_thermo.yaml.Check spelling and composition.
Species C2H2+ not found in grc_nasa9_all_C_H_species.yaml.Check spelling and composition.
Species C2H2+ not found in mutationpp_nasa9.yaml.Check spelling and composition.
Species C2H2+ not found in nasa1993detchem2005_298-5000K_thermo.yaml.Check spelling and composition.
Species H2+ not found in burcat_thermo_nasa9.yaml.Check spelling and composition.
Total running time of the script: (0 minutes 2.355 seconds)